Kocuria spp. in Foods: Biotechnological Uses and Risks for Food Safety
Applied Food Biotechnology,
Vol. 8 No. 2 (2021),
16 March 2021
,
Page 79-88
https://doi.org/10.22037/afb.v8i2.30748
Abstract
.png)
Background and objective:
Bacteria of the Genus Kocuria are found in several environments and their isolation from foods has recently increased due to more precise identification protocols using molecular and instrumental techniques. This review describes biotechnological properties and food-linked aspects of these bacteria, which are closely associated with clinical cases.
Results and conclusion:
Kocuria spp. are capable of production of various enzymes, being potentially used in environmental treatment processes and clinics and production of antimicrobial substances. Furthermore, these bacteria show desirable enzymatic activities in foods such as production of catalases and proteases. Beneficial interactions with other microorganisms have been reported on increased production of enzymes and volatile compounds in foods. However, there are concerns about the bacteria, including their biofilm production, which generates technological and safety problems. The bacterial resistance to antimicrobials is another concern since isolates of this genus are often resistant or multi-resistant to antimicrobials, which increases the risk of gene transfer to pathogens of foods.
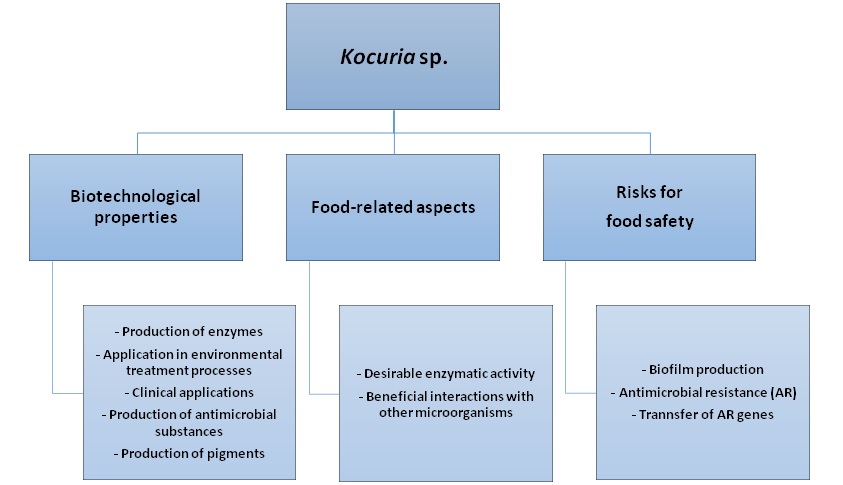
- Kocuria spp.
- Gram-positive cocci
- Biotechnological potential
- Biofilm
- Antimicrobial resistance.
How to Cite
References
Stackebrandt E, Koch C, Gvozdiak O, Schumann P. Taxonomic dissection of the genus Micrococcus: Kocuria gen. nov., Nesterenkonia gen. nov., Kytococcus gen. nov., Dermacoccus gen. nov., and Micrococcus Cohn 1872 gen. emend. Int J Syst Evol Micr. 1995; 45 (4): 682-692.
doi: 10.1099/00207713-45-4-682
Li J, Zhang S. Kocuria coralli sp. nov., a novel actinobacterium isolated from coral reef seawater. Int J Syst Evol Micr. 2020; 70 (2): 785-789.
doi: 10.1099/ijsem.0.003825
LPSN. List of Prokaryotic names with Standing in Nomenclature. Available from:
https://lpsn.dsmz.de/genus/kocuria [Accessed 15 May 2020].
Kandi V, Palange P, Vaish R, Bhatti AB, Kale V, Kandi MR, Bhoomagiri MR. Emerging bacterial infection: Identification and clinical significance of Kocuria species. Cureus 2016; 8 (8): e731.
doi: /10.7759/cureus.731
Braun MS, Wang E, Zimmermann S, Boutin S, Wink M. Kocuria uropygioeca sp. nov. And Kocuria uropygialis sp. nov., isolated from the preen glands of Great Spotted Woodpeckers (Dendrocopos major). Syst Appl Microbiol. 2018; 41 (1): 38-43.
doi: 10.1016/j.syapm.2017.09.005
Centeno JA, Garabal JI, Docampo F, Lorenzo JM, Carballo J. Recovering traditional raw-milk Tetilla cheese flavour and sensory attributes by using Kocuria varians and Yarrowia lipolytica adjunct cultures. Int J Food Microbiol. 2017; 251: 33-40.
doi: 10.1016/j.ijfoodmicro.2017.03.014
Organji SR, Abulreesh HH, Elbanna K, Osman GE, Almalki MH. Diversity and characterization of Staphylococcus spp. in food and dairy products: A foodstuff safety assessment. J Microbiol Biotechnol Food Sci. 2018; 7 (6): 586.
doi: 10.15414/jmbfs.2018.7.6.586-593
Machado MAA, Ribeiro WA, Toledo VS, Ramos GLPA, Vigoder HC, Nascimento JS. Antibiotic resistance and biofilm production in Gram-positive catalase-positive cocci isolated from Brazilian pasteurized milk. J. Food Qual. Hazards Control. 2020; 7: 67-74.
doi: 10.18502/jfqhc.7.2.2886
Uzair B, Menaa F, Khan BA, Mohammad FV, Ahmad VU, Djeribi R, Menaa B. Isolation, purification, structural elucidation and antimicrobial activities of kocumarin, a novel antibiotic isolated from actinobacterium Kocuria marina CMG S2 associated with the brown seaweed Pelvetia canaliculata. Microbiol. Res. 2018; 206: 186-197.
doi: 10.1016/j.micres.2017.10.007
Gardini F, Tofalo R, Suzzi G. A survey of antibiotic resistance in Micrococcaceae isolated from Italian dry fermented sausages. J Food Protect. 2003; 66 (6): 937-945.
doi: 10.4315/0362-028x-66.6.937
Tremonte P, Succi M, Di Renzo T, Reale A, Sorrentino E, Coppola R. Interactions between strains of Staphylococcus xylosus and Kocuria varians isolated from fermented meats. J Appl Microbiol. 2007; 103: 743-751.
doi: 10.1111/j.1365-2672.2007.03315.x
Noor-Uddin GM, Larsen MH, Guardabassi L, Dalsgaard A. Bacterial flora and antimicrobial resistance in raw frozen cultured seafood imported to Denmark. J Food Protect. 2013; 76 (3): 490-499.
doi: 10.4315/0362-028x.jfp-12-402
Boruah P, Sarmah P, Das PK, Goswami, T. Exploring the lignolytic potential of a new laccase producing strain Kocuria sp. PBS-1 and its application in bamboo pulp bleaching. Int Biodeter Biodegrad. 2019; 143: 104726.
doi: 10.1016/j.ibiod.2019.104726
Fancello F, Multineddu C, Santona M, Deiana P, Zara G, Mannazzu I, Budroni M, Dettori S, Zara S. Bacterial biodiversity of extra virgin olive oils and their potential biotechnological exploitation. Microorganisms 2020; 8 (1): 97.
doi: 10.3390/microorganisms8010097
Kacaniova M, Gasper J, Brindza J, Schubertova Z, Ivanisova E. Bacteria of apis mellifera gastrointestinal tract: Counts, identification and their antibiotic resistance. Agrobiodivers Improv Nutr Health Life Qual. 2017; 1: 210-215
doi: 10.15414/agrobiodiversity.2017.2585-8246.210-215
Gagnon M, Hamelin L, Frechette A, Dufour S, Roy D. Effect of recycled manure solids as bedding on bulk tank milk and implications for cheese microbiological quality. J Dairy Sci. 2019; 103 (1):128-140.
doi: 10.3168/jds.2019-16812
Purty S, Saranathan R, Prashanth K, Narayanan K, Asir J, Sheela Devi C, Amarnath SK. The expanding spectrum of human infections caused by Kocuria species: A case report and literature review. Emerg Microbes Infect. 2013; 2 (1): 1-8.
doi: 10.1038/emi.2013.93
Dotis J, Printza N, Stabouli S, Papachristou F. Kocuria species periotonitis: Although rare, we have to care. Periton. Dialysis Int. 2015; 35 (1): 26-30.
doi: 10.3747/pdi.2013.00138
Savini V, Catavitello C, Masciarelli G, Astolfi D, Babinot A, Bianco A, Febbo F, DAmario C, DAntonio D. Drug sensitivity and clinical impact of members of the genus Kocuria. J Med Microbiol. 2010; 59: 1395-1402.
doi: 10.1099/jmm.0.021709-0
Reddy GS, Prakash JS, Prabahar V, Matsumoto GI, Stackebrandt E, Shivaji S. Kocuria polaris sp. nov., an orange-pigmented psychrophilic bacterium isolated from an Antarctic cyanobacterial mat sample. Int J Syst Evol Micr. 2003; 53 (1): 183-187.
doi:10.1099/ijs.0.02336-0
Bertsch A, Coello NA. Biotechnological process for treatment and recycling poultry feathers as a feed ingredient. Bioresource Technol. 2005; 96 (3): 1703-1708.
doi: 10.1016/j.biortech.2004.12.026
Rezaeeyan Z, Safarpour A, Amoozegar MA, Babavalian H, Tebyanian H, Shakeri F. High carotenoid production by a halotolerant bacterium, Kocuria sp. strain QWT-12 and anticancer activity of its carotenoid. Excli J. 2017; 16: 840-851.
doi: 10.17179/excli2017-218
Bernal C, Vidal L, Valdivieso E, Coello, N. Keratinolytic activity of Kocuria rosea. World J Microb Biot. 2003; 19: 255-261.
doi: 10.1023/A:1023685621215
Lalevic B, Raicevic V, Atanaskovic I, Kikovic D, Talai-ekhozani A, Hamidovic S, Gkorezis P. Biodegradation of crude oil by Kocuria sp. J Environ Treat Tech. 2014; 2 (3): 99-101.
El Mahdi AM, Aziz HA, Abu Am SS, El-Gendy NS, Nassar H. Performance of isolated Kocuria sp. SAR1 in light crude oil biodegradation. J Bioremediat Biodegrad. 2015; 6: 303.
doi: 10.4172/2155-6199.1000303
Haq F, Butt M, Ali H, Chaudhary HJ. Biosorption of cadmium and chromium from water by endophytic Kocuria rhizophila: Equilibrium and kinetic studies. Desalin Water Treat. 2016; 57 (42): 19946-19958.
doi: 10.1080/19443994.2015.1109561
Anum S, Khan SM, Chaudhary HJ, Ahmad Z, Afza R. Phytoremediation of nickel polluted ecosystem through selected ornamental plant species in the presence of bacterium Kocuria rhizophila. Bioremediat J. 2019; 23 (3): 215-226.
doi: 10.1080/10889868.2019.1639610
Zhou L, Dong F, Li H, Huo T, Wang P, Liu M, He H. Surface interaction between metazeunerite and an indigenous micro-organism Kocuria rosea: Implications for bioremediation of as-U tailings. Chem Eng J. 2019; 359: 393-401.
doi: 10.1016/j.cej.2018.11.154
Drosinos EH, Paramithiotis S, Kolovos G, Tsikouras I, Metaxopoulos I. Phenotypic and technological diversity of lactic acid bacteria and staphylococci isolated from traditionally fermented sausages in southern Greece. Food Microbiol. 2007; 24 (3):260-270.
doi: 10.1016/j.fm.2006.05.001
Kamiloglu A, Kaban G, Kaya M. Contribution of catalase positive cocci on flavour formation in fermented sausages. Curr J Appl Sci Technol. 2016; 17 (1):1-8.
doi: 10.1016/j.jbiotec.2014.07.085
Laranjo M, Elias M, Fraqueza MJ. The use of starter cultures in traditional meat products. J Food Qual. 2017.
doi: 10.1155/2017/9546026
Cruxen CES, Funck GD, Haubert L, Dannenberg GS, Marques JL, Chaves FC, Silva WP, Fiorentini AM. Selection of native bacterial starter culture in the production of fermented meat sausages: Application potential, safety aspects, and emerging technologies. Food Res Int. 2019; 122: 371-382.
doi: 10.1016/j.foodres.2019.04.018
Feng Z, Chen X, Li J, Ren D. An alkaline protease from Kocuria kristinae F7: properties and characterization of its hydrolysates from soy protein. Eur Food Res Technol. 2013; 236: 293-301.
doi: 10.1007/s00217-012-1890-9
Feng Z, Chen H, Lv XT, Deng HL, Chen X, Li JJ, Guo L. Accelerated ripening of kedong sufu with autochthonous starter cultures Kocuria rosea KDF3 and its protease KP3 as adjuncts. J Appl Microbiol. 2014; 116 (4): 877-889.
doi: 10.1111/jam.12433
Callejon S, Sendra R, Ferrer S, Pardo I. Ability of Kocuria varians LTH 1540 to degrade putrescine: Identification and characterization of a novel amine oxidase. J Agr Food Chem. 2015; 63 (16): 4170-4178.
doi: 10.1021/jf5026967
Pridmore D, Rekhif N, Pittet AC, Suri B, Mollet B. Variacin, a new lanthionine-containing bacteriocin produced by Micro-coccus varians: Comparison to lacticin 481 of Lactococcus lactis. Appl Environ Microbiol. 1996; 62 (5): 1799-1802.
doi: 10.1128/AEM.62.5.1799-1802.1996
O’Mahony T, Rekhif N, Cavadini C, Fitzgerald GF. The application of a fermented food ingredient containing ‘variacin’, a novel antimicrobial produced by Kocuria varians, to control the growth of Bacillus cereus in chilled dairy products. J Appl Microbiol. 2001; 90: 106-114.
doi: 10.1046/j.1365-2672.2001.01222.x
Martin J, Sousa ST, Crespo G, Palomo S, Gonzalez I, Tormo J, Cruz M, Anderson M, Hill R, Vicente F, Genilloud O, Reyes F. Kocurin, the true structure of PM181104, an anti-methicillin-resistant Staphylococcus aureus (MRSA) thiazolyl peptide from the marine-derived bacterium Kocuria palustris. Mar Drugs. 2013; 11 (12): 387-398.
doi: 10.3390/md11020387
Bundale S, Singh J, Begde D, Nashikkar N, Upadhyay A. Rare actinobacteria: a potential source of bioactive polyketides and peptides. World J Microbiol Biotechnol. 2019; 35 (6): 92.
doi: 10.1007/s11274-019-2668-z
Ibarra-Sanchez LA, El-Haddad N, Mahmoud D, Miller MJ, Karam L. Invited review: Advances in nisin use for preservation of dairy products. J Dairy Sci. 2020; 103 (3): 2041-2052.
doi: 10.3168/jds.2019-17498
Donlan RM. Biofilms: Microbial life on surfaces. Emerg Infect Dis. 2002; 8 (9): 881-890.
doi: 10.3201/eid0809.020063
Weber M, Liedtke J, Plattes S, Lipski A. Bacterial community composition of biofilms in milking machines of two dairy farms assessed by a combination of culture-dependent and-independent methods. Plos One 2019; 14 (9): e0222238.
doi: 10.1371/journal.pone.0222238
Teh A, Lee S, Dykes G. Does Campylobacter jejuni form biofilms in food-related environments?. Appl Environ Microbiol. 2014; 80 (17): 5154-5160.
doi: 10.1128/aem.01493-14
Bridier A, Sanchez-Vizuete P, Guilbaud M, Piard JC, Naitali M, Briandet R. Biofilm-associated persistence of food-borne pathogens. Food Microbiol. 2015; 45: 67-178.
doi: 10.1016/j.fm.2014.04.015
Carpentier B, Chassaing D. Interactions in biofilms between Listeria monocytogenes and resident microorganisms from food industry premises. Int J Food Microbiol. 2004; 97: 111-122.
doi: 10.1016/j.ijfoodmicro.2004.03.031
Midelet G, Kobilinsky A, Carpentier B. Construction and analysis of fractional multifactorial designs to study attachment strength and transfer of Listeria monocytogenes from pure or mixed biofilms after contact with a solid model food. Appl Environ Microbiol. 2006; 72 (4): 2313-2321.
doi: 10.1128/AEM.72.4.2313-2321.2006
Roder HL, Raghupathi PK, Herschend J, Brejnrod A, Knochel S, Sorensen SJ, Burmolle M. Interspecies interactions result in enhanced biofilm formation by co-cultures of bacteria isolated from a food processing environment. Food Microbiol. 2015; 51: 18-24.
doi: 10.1016/j.fm.2015.04.008
Lim ES, Lee JE, Kim JS, Koo OK. Isolation of indigenous bacteria from a cafeteria kitchen and their biofilm formation and disinfectant susceptibility. LWT 2017; 77: 376-382.
doi: 10.1016/j.lwt.2016.11.060
Leriche V, Carpentier B. Limitation of adhesion and growth of Listeria monocytogenes on stainless steel surfaces by Staphylococcus sciuri biofilms. J Appl Microbiol 2000; 88 (4): 594-605.
doi: 10.1046/j.1365-2672.2000.01000.x
Argudin M, Deplano A, Meghraoui A, Dodemont M, Heinrichs A, Denis O, Nonhoff C, Roisin S. Bacteria from animals as a pool of antimicrobial resistance genes. Antibiot. 2017; 6 (2): 12.
doi: 10.3390/antibiotics6020012
Toth AG, Csabai I, Kriko E, Tozser D, Maroti G, Patai AV, Makrai L, Szita G, Solymosi N. Raw milk for human consumption may carry antimicrobial resistance genes. Biorxiv. 2019; 853333: 2-7.
doi: 10.1101/853333
Toth AG, Csabai I, Kriko E, Tozser D, Maroti G, Patai AV, Makrai L, Szita G, Solymosi N. Antimicrobial resistance genes in raw milk for human consumption. Sci Rep. 2020; 10: 7464.
doi: 10.1038/s41598-020-63675-4
Perrin-Guyomard A, Soumet C, Leclercq R, Doucet-Populaire F, Sanders P. Antibiotic susceptibility of bacteria isolated from pasteurized milk and characterization of macrolide-lincos-amide-streptogramin resistance genes. J Food Protect. 2005; 68 (2): 347-352.
doi: 10.4315/0362-028x-68.2.347
Pekala A, Pazdzior E, Antychowicz J, Bernad A, Głowacka H, Wiecek B, Niemczuk W. Kocuria rhizophila and Micrococcus luteus as emerging opportunist pathogens in brown trout (Salmo trutta Linnaeus, 1758) and rainbow trout (Oncorhy-nchus mykiss Walbaum, 1792). Aquacult. 2018; 486: 285-289.
doi: 10.1016/j.aquaculture.2017.12.028
Rodriguez-Alonso P, Fernandez-Otero C, Centeno JA, Garabal JI. Antibiotic resistance in lactic acid bacteria and Microco-ccaceael Staphylococcaceae isolates from artisanal raw milk cheeses, and potential implications on cheese making. J Food Sci. 2009; 74 (6): M284-M293.
doi: 10.1111/j.1750-3841.2009.01217.x
Cook PW, Nightingale KK. Use of omics methods for the advancement of food quality and food safety. Anim Front. 2018; 8(4): 33-41.
doi:10.1093/af/vfy024
Bergholz TM, Moreno Switt AI, Wiedmann M. Omics approaches in food safety: Fulfilling the promise?. Trends Microbiol. 2014; 22(5): 275-281.
doi:10.1016/j.tim.2014.01.006
Haddad N, Johnson N, Kathariou S, Metris A, Phister T, Pielaat A, Tassou C, Wells-Benik MHJ, Zwietering MH. Next generation microbiological risk assessment-Potential of omics data for hazard characterisation. Int J Food Microbiol. 2018; 287(20): 28-39.
doi:10.1016/j.ijfoodmicro.2018.04.015
O’Flaherty S, Klaenhammer TR. The impact of omic technologies on the study of food microbes. Annu Rev Food Sci Technol. 2011; 2(1): 353-371.
doi:10.1146/annurev-food-030810-110338
Bridier A. Exploring Foodborne Pathogen Ecology and Antimicrobial Resistance in the Light of Shotgun Metagenomics. In: Bridier A. (Eds.) Foodborne Bacterial Pathogens. Methods in Molecular Biology. Humana, New York, NY. 2018: pp.229-245.
doi: 10.1007/978-1-4939-9000-9_19
Codex Alimentarius Commission. Procedural Manual of the Codex Alimentarius Commission. 26th edition. 2018. Available from: http://www.fao.org/3/i8608en/i8608en.pdf
Franz CM, den Besten, HM, Boehnlein C, Gareis M, Zwietering MH, Fusco V. Reprint of: Microbial food safety in the 21st century: Emerging challenges and foodborne pathogenic bacteria. Trends Food Sci Technol. 2019;84:34-37.
doi: 10.1016/j.tifs.2019.01.009
- Abstract Viewed: 1214 times
- pdf Downloaded: 982 times
