Overview of Catalytic Properties of Fungal Xylose Reductases and Molecular Engineering Approaches for Improved Xylose Utilisation in Yeast
Applied Food Biotechnology,
Vol. 5 No. 2 (2018),
23 March 2018
,
Page 47-58
https://doi.org/10.22037/afb.v5i2.19233
Abstract
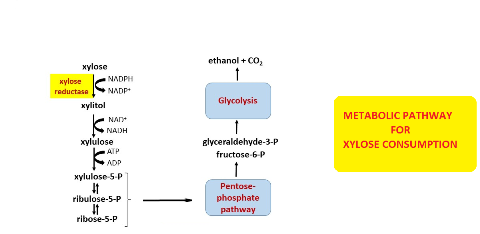
Background and Objective: Xylose reductases belong to the aldo-keto reductase family of enzymes, which catalyse the conversion of xylose to xylitol. Yeast xylose reductases have been intensively studied in the last two decades due to their significance in biotechnological production of ethanol and xylitol from xylose. Due to its GRAS status and pronounced tolerance to harsh conditions, Saccharomyces cerevisiae is the ideal organism for industrial production of both xylitol and ethanol. However, Saccharomyces cerevisiae is unable to use xylose as the sole carbon source due to the lack of xylose specific transporters and insufficient activity of metabolic pathways for xylose utilisation. The aim of this paper is to give an overview of attempts in increasing biotechnological potential of xylose reductases and to highlight the prospective of this application.
Results and Conclusion: In order to create strains with improved xylose utilization, different approaches were attempted including simultaneous overexpression of xylitol dehydrogenase, xylose reductase and pentose phosphate pathway enzymes, heterologous expression of putative xylose transporters or heterologous expression of genes coding for enzymes included in the xylose metabolism, respectively. Furthermore, number of attempts to genetically modify different xylose reductases is increasing. This review presents current knowledge about yeast xylose reductases and the different approaches applied in order to improve xylose metabolism in yeast.
Conflict of interest: The authors declare no conflict of interest.- ▪ Fungal xylose reductase ▪ Xylitol ▪ Xylose bioconversion ▪ Xylose metabolism
How to Cite
References
Salusjärvi L, Kankainen M, Soliymani R, Pitkanen JP, Penttila M, Ruohonen L. Regulation of xylose metabolism in recombinant Saccharomyces cerevisiae. Microb Cell Fact. 2008; 7:18. doi: 10.1186/1475-2859-7-18
Farrell AE, Plevin RJ, Turner BT, Jones AD, O’Hare M, Kammen DM. Ethanol can contribute to energy and environmental goals. Science. 2006; 311(5760):506-508. doi: 10.1126/science.1121416
van Maris AJA, Abbott DA, Bellissimi E, van den Brink J, Kuyper M, Luttik MA, Wisselink HW, Scheffers WA, van Dijken JP, Pronk JT. Alcoholic fermentation of carbon sources in biomass hydrolysates by Saccharomyces cerevisiae: Current status. Antonie Van Leeuwenhoek. 2006; 90(4):391-418. doi: 10.1007/s10482-006-9085-7
Pauly M, Keegstra K. Cell-wall carbohydrates and their modification as a resource for biofuels. Plant J. 2008; 54(4):559-568. doi: 10.1111/j.1365-313X.2008.03463.x
Runquist D, Hahn-Hagerdal B, Radstrom P. Comparison of heterologous xylose transporters in recombinant Saccharomyces cerevisiae. Biotechnol Biofuels. 2010; 3:5. doi: 10.1186/1754-6834-3-5
Young EM, Tong A, Bui H, Spofford C, Alper HS. Rewiring yeast sugar transporter preference through modifying a conserved protein motif. Proc Natl Acad Sci USA. 2014; 111(1):131-136. doi: 10.1073/pnas.1311970111
Bera AK, Ho NW, Khan A, Sedlak M. A genetic overhaul of Saccharomyces cerevisiae 424A(LNH-ST) to improve xylose fermentation. J Ind Microbiol Biotechnol. 2011; 38(5):617-626. doi: 10.1007/s10295-010-0806-6
Demeke MM, Dietz H, Li Y, Foulquie-Moreno MR, Mutturi S, Deprez S, Den Abt T, Bonini BM, Liden G, Dumortier F, Verplaetse A, Boles E, Thevelein JM. Development of a D-xylose fermenting and inhibitor tolerant industrial Saccharomyces cerevisiae strain with high performance in lignocellulose hydrolysates using metabolic and evolutionary engineering. Biotechnol Biofuels. 2013; 6(1):89. doi: 10.1186/1754-6834-6-89
Toivari MH, Salusjarvi L, Ruohonen L, Penttilä M. Endogenous xylose pathway in Saccharomyces cerevisiae. J Appl Environ Microbiol. 2004; 70(6):3681-3686. doi: 10.1128/AEM.70.6.3681-3686.2004
Hahn-Hagerdal B, Karhumaa K, Fonseca C, Spencer-Martins I, Gorwa-Grauslund MF. Towards industrial pentose-fermenting yeast strains. J Appl Microbiol Biotechnol. 2007; 74(5):937-953. doi: 10.1007/s00253-006-0827-2
Kim SR, Park YC, Jin YS, Seo JH. Strain engineering of Saccharomyces cerevisiae for enhanced xylose metabolism. Biotechnol Adv. 2013; 31(6):851-861. doi: 10.1016/j.biotechadv.2013.03.004
van Maris AJ, Winkler AA, Kuyper M, de Laat WT, van Dijken JP, Pronk JT. Development of efficient xylose fermentation in Saccharomyces cerevisiae: xylose isomerase as a key component. Adv Biochem Eng Biotechnol. 2007; 108:179-204. doi: 10.1007/10_2007_057
Bengtsson O, Hahn-Hägerdal B, Gorwa-Grauslund MF. Xylose reductase from Pichia stipitis with altered coenzyme preference improves ethanolic xylose fermentation by recombinant Saccharomyces cerevisiae. Biotechnol Biofuels. 2009; 2:9. doi: 10.1186/1754-6834-2-9
Dmytruk OV, Dmytruk KV, Abbas CA, Voronovsky AY, Sibirny AA. Engineering of xylose reductase and overexpression of xylitol dehydrogenase and xylulokinase improves xylose alcoholic fermentation in the thermotolerant yeast Hansenula polymorpha. Microb Cell Fact. 2008; 7:21. doi: 10.1186/1475-2859-7-21
Liang L, Zhang J, Lin Z. Altering coenzyme specificity of Pichia stipitis xylose reductase by the semi-rational approach CASTing. Microb Cell Fact. 2007; 6:36. doi: 10.1186/1475-2859-6-36
Petschacher B, Leitgeb S, Kavanagh KL, Wilson DK, Nidetzky B. The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and X-ray crystallography. J Biochem. 2005; 385(1):75-83. doi: 10.1042/BJ20040363
Runquist D, Hahn-Hagerdal B, Bettiga M. Increased Ethanol Productivity in Xylose-Utilizing Saccharomyces cerevisiae via a Randomly Mutagenized Xylose Reductase. J Appl Environ Microbiol. 2010a; 76(23):7796-7802. doi: 10.1128/AEM.01505-10
Watanabe S, Saleh AA, Pack SP, Annaluru N, Kodaki T, Makino K. Ethanol production from xylose by recombinant Saccharomyces cerevisiae expressing protein engineered NADH-preferring xylose reductase from Pichia stipitis. Microbiology. 2007; 153(9):3044-3054.
doi: 10.1099/mic.0.2007/007856-0
Diao L, Liu Y, Qian F, Yang J, Jiang Y, Yang S. Construction of fast xylose fermenting yeast based on industrial ethanol-producing diploid Saccharomyces cerevisiae by rational design and adaptive evolution. BMC Biotechnol. 2013; 13:110. doi: 10.1186/1472-6750-13-110
Scalcinati G, Otero JM, Van Vleet JR, Jeffries TW, Olsson L, Nielsen J. Evolutionary engineering of Saccharomyces cerevisiae for efficient aerobic xylose consumption. FEMS Yeast Res. 2012; 12(5):582-597. doi: 10.1111/j.1567-1364.2012.00808.x
Kim SR, Skerker JM, Kang W, Lesmana A, Wei N, Arkin AP, Jin YS. Rational and evolutionary engineering approaches uncover a small set of genetic changes efficient for rapid xylose fermentation in Saccharomyces cerevisiae. PLoS One. 2013a; 8(2):e57048. doi: 10.1371/journal.pone.0057048
Ko BS, Kim J, Kim JH. Production of Xylitol from D-Xylose by a Xylitol Dehydrogenase Gene-Disrupted Mutant of Candida tropicalis. J Appl Environ Microbiol. 2006; 72(6):4207-4213. doi: 10.1128/AEM.02699-05
Wang TH, Zhong YH, Huang W, Liu T, You YW. Antisense inhibition of xylitol dehydrogenase gene, xdh1 from Trichoderma reesei. Lett Appl Microbiol. 2005; 40(6):424-429. doi: 10.1111/j.1472-765X.2005.01685.x
Prakasham RS, Rao RS, Hobbs PJ. Current trends in Biotechnological Production of Xylitol and Future Prospects. Curr Trends Biotechnol Pharm. 2009; 3(1):8-36
Hagedorn J, Ciriacy M. Isolation and characterization of xyl mutants in a xylose-utilizing yeast, Pichia stipitis. Curr Genet. 1989; 16(1):27-33. doi: 10.1007/BF00411080
Bolen PL, Roth KA, Freer SN. Affinity Purifications of Aldose Reductase and Xylitol Dehydrogenase from the Xylose-Fermenting Yeast Pachysolen tannophilus. J Appl Environ Microbiol. 1986; 52(4):660-664
Chu BCH, Lee H. Investigation of the Role of a Conserved Glycine Motif in the Saccharomyces cerevisiae Xylose Reductase. Curr Microbiol. 2006; 53(2):118-123. doi: 10.1007/s00284-005-0325-2
Cocotle-Ronzon Y, Zendejas-Zaldo M, López del Castillo-Lozano M, Aguilar-Uscanga M. Preliminary Characterization of Xylose Reductase Partially Purified by Reversed Micelles from Candida tropicalis IEC5-ITV, an Indigenous Xylitol-Producing Strain. Advances in Chemical Engineering and Science. 2012; 2(1):9-14. doi: 10.4236/aces.2012.21002
Cortez EV, Pessoa Jr. A, Almeida-Felipe MG, Roberto IC and Vitolo M. Characterization of Xylose Reductase Extracted by CTAB-Reversed Micelles from Candida guilliermondii Homogenate. Braz J Pharm Sci. 2006; 42 (2):251-257. doi: 10.1590/S1516-93322006000200010
Ditzelmüller G, Kubicek CP, Wöhrer W, Röhr M. Xylose metabolism in Pachysolen tannophilus: purification and properties of xylose reductase. Can J Microbiol. 1984; 30(11): 1330-1336. doi: 10.1139/m84-214
Ditzelmüller G, Kubicek-Pranz EM, Röhr M, Kubicek CP. NADPH-specific and NADH-specific xylose reduction is catalyzed by two separate enzymes in Pachysolen tannophilus. J Appl Microbiol Biotechnol. 1985; 22(4):297-299. doi: 10.1007/BF00252033
Fernandes S, Tuohy MG, Murray PG. Xylose reductase from the thermophilic fungus Talaromyces emersonii: cloning and heterologous expression of the native gene (Texr) and a double mutant (TexrK271R + N273D) with altered coenzyme specificity. J Biosci. 2009; 34(6):881-890.
doi: 10.1007/s12038-009-0102-7
Jeong EY, Kim IS, Lee H. Identification of lysine-78 as an essential residue in the Saccharomyces cerevisiae xylose reductase. FEMS Microbiol Lett. 2002; 209(2):223-228. doi: 10.1111/j.1574-6968.2002.tb11135.x
Kavanagh KL, Klimacek M, Nidetzky B, Wilson DK. The structure of apo and holo forms of xylose reductase, a dimeric aldoketo reductase from Candida tenuis. Biochemistry. 2002; 41(28):8785-8795. doi: 10.1021/bi025786n
Kavanagh KL, Klimacek M, Nidetzky B, Wilson DK. Structure of xylose reductase bound to NAD+ and the basis for single and dual co-substrate specificity in family 2 aldo-keto reductases. Biochem J. 2003; 373(2):319-326. doi: 10.1042/BJ20030286
Kuhn A, van Zyl C, van Tonder A, Prior BA. Purification and Partial Characterization of an Aldo-Keto Reductase from Saccharomyces cerevisiae. J Appl Environ Microbiol. 1995; 61(4):1580-1585
Kumar S, Gummadi SN. Purification and biochemical characterization of a moderately halotolerant NADPH dependent xylose reductase from Debaryomyces nepalensis NCYC 3413. Bioresour Technol. 2011; 102(20):9710-9717. doi: 10.1016/j.biortech.2011.07.030
Lee JK, Koo BS, Kim SY. Cloning and characterization of the xyl1 gene, encoding an NADH-preferring xylose reductase from Candida parapsilosis, and its functional expression in Candida tropicalis. J Appl Environ Microbiol. 2003; 69(10):6179-6188. doi: 10.1128/AEM.69.10.6179-6188.2003
Mayr P, Bruggler K, Kulbe KD, Nidetzky B. D-Xylose metabolism by Candida intermedia: isolation and characterisation of two forms of aldose reductase with different coenzyme specificities. J Chromatogr. 2000; 737(1-2):195-202. doi: 10.1016/S0378-4347(99)00380-1
Morimoto S, Tawaratani T, Azuma K, Oshima T, Sinskey AJ. Purification and properties of aldose reductase form Pachysolen tannophilus. J Ferment Technol. 1987; 65(1):17-21. doi: 10.1016/0385-6380(87)90060-4
Neuhauser W, Haltrich D, Kulbe KD, Nidetzky B. NAD(P)H dependent aldose reductase from the xylose-assimilating yeast Candida tenuis. Isolation, characterization and biochemical properties of the enzyme. Biochem J. 1997; 326(3):683-692
Nolleau V, Preziosi-Belloy L, Delgenes JP, Navarro JM. Xylitol production from xylose by two yeast strains: Sugar tolerance. Curr Microbiol. 1993; 27(4):191-197. doi: 10.1007/BF01692875
Sampaio FC, de Faria JT, Lopes Passos FM, Converti A, Minin LA. Optimal activity and thermostability of xylose reductase from Debaryomyces hansenii UFV-170. J Ind Microbiol Biotechnol. 2009; 36(2):293-300. doi: 10.1007/s10295-008-0498-3
Sene L, Felipe MGA, Silva SS, Vitolo M. Preliminary kinetic characterization of xylose reductase and xylitol dehydrogenase extracted from Candida guilliermondii FTI 20037 cultivated in sugarcane bagasse hydrolysate for xylitol production. Biotechnol Appl Biochem. 2001; 91(1-9):671-680. doi: 10.1385/ABAB:91-93:1-9:671
Silva SS, Vitolo M, Pessoa Jr. A, Felipe MGA. Xylose reductase and xylitol dehydrogenase activities of D-xylose-xylitol-fermenting Candida guilliermondii. J Basic Microbiol. 1996; 36(3):187-191. doi: 10.1002/jobm.3620360305
Träff KL, Jönsson LJ, Hahn-Hägerdal B. Putative xylose and arabinose reductases in Saccharomyces cerevisiae. Yeast. 2002; 19(14): 1233-1241. doi: 10.1002/yea.913
Verduyn C, Vankleef R, Frank J, Schreuder H, Vandijken JP, Scheffers WA. Properties of the NAD(P)H-dependent xylose reductase from the xylose-fermenting yeast Pichia stipitis. Biochem J. 1985; 226(3):669-677. doi: 10.1042/bj2260669
Woodyer R, Simurdiak M, van der Donk WA, Zhao H. Heterologous Expression, Purification, and Characterization of a Highly Active Xylose Reductase from Neurospora crasa. J Appl Environ Microbiol. 2005; 71(3):1642-1647. doi: 10.1128/AEM.71.3.1642-1647.2005
Yokoyama S, Suzuki T, Kawai K, Horitsu H, Takamizawa K. Purification, characterization and structure analysis of NADPH dependent D-xylose reductases from Candida tropicalis. J Ferment Bioeng. 1995; 79(3):217-223. doi: 10.1016/0922-338X(95)90606-Z
Zhang M, Jiang S, Zheng Z, Li X, Luo S, Wu X. Cloning, expression, and characterization of a novel xylose reductase from Rhizopus oryzae. J Basic Microbiol. 2015; 55(7):907-921. doi: 10.1002/jobm.201400786
Ellis EM. Microbial aldo-keto reductases. FEMS Microbiol Lett. 2002; 216(2):123-131. doi: 10.1111/j.1574-6968.2002.tb11425.x
Nidetzky B, Bruggler K, Kratzer R, Mayr P. Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using quantitative structure-activity relationships, steady-state kinetic analysis, and pH studies. J Agric Food Chem. 2003; 51(27):7930-7935. doi: 10.1021/jf034426j
Amore R, Kotter P, Kuster C, Ciriacy M, Hollenberg CP. Cloning and expression in Saccharomyces cerevisiae of the NAD(P)H-dependent xylose reductase-encoding gene (Xyl1) from the xylose-assimilating yeast Pichia stipitis. Gene. 1991; 109(1):89-97. doi: 10.1016/0378-1119(91)90592-Y
Hacker B, Habenicht A, Kiess M, Mattes R. Xylose utilisation: cloning and characterisation of the xylose reductase from Candida tenuis. Biol Chem. 1999; 380(12):1395-1403. doi: 10.1515/BC.1999.179
Kang MH, Ni H, Jeffries TW. Molecular characterization of a gene for aldose reductase (CbXYL1) from Candida boidinii and its expression in Saccharomyces cerevisiae. Biotechnol Appl Biochem. 2003; 105-108:265–276
Schneider H, Lee H, Barbosa M de FS, Kubicek CP, James AP. Physiological properties of a mutant of Pachysolen tannophilus deficient in NADPH-dependent D-xylose reductase. J Appl Environ Microbiol. 1989; 55(11):2877-2881
Webb SR, Lee H. Inhibitors of xylose reductase from the yeast Pichia stipitis. Biotechnol Appl Biochem. 1991; 30:325-337. doi: 10.1007/BF02922036
Webb SR, Lee H. Characterization of xylose reductase from the yeast Pichia stipitis: evidence for functional thiol and histidyl groups. J Gen Microbiol. 1992; 138(9):1857-1863. doi: 10.1099/00221287-138-9-1857
Jörnvall H, Persson B, Krook M, Atrian S, Gonzalez-Duarte R, Jeffery J, Ghosh D. Short-chain dehydrogenases/reductases (SDR). Biochemistry. 1995; 34(18):6003-6013. doi: 10.1021/bi00018a001
Lee H. The Structure and Function of Yeast Xylose (Aldose) Reductases. Yeast. 1998; 14(11):977-984. doi: 10.1002/(SICI)1097-0061(199808)14:11<977::AID-YEA302>3.0.CO;2-J
Rizzi M, Erlemann P, Bui-Thanh NA, Dellweg H. Xylose fermentation by yeasts. 4. Purification and kinetic studies of xylose reductase from Pichia stipitis. J Appl Microbiol Biotechnol. 1988; 29(2-3):148-154. doi: 10.1007/BF00939299
Bruinenberg PM, de But PHM, van Dijken JP, Scheffers WA. NADH-linked aldose reductase: the key to anaerobic alcoholic fermentation of xylose by yeasts. J Appl Microbiol Biotechnol. 1984; 19(4):256-260. doi: 10.1007/BF00251847
Zhang Y, Lee H. Site-directed mutagenesis of the cysteine residues in the Pichia stipitis xylose reductase. FEMS Microbiol Lett. 1997; 147(2):227-232. doi: 10.1111/j.1574-6968.1997.tb10246.x
Kostrzynska M, Sopher CR, Lee H. Mutational analysis of the role of the conserved lysine-270 in the Pichia stipitis xylose reductase. FEMS Microbiol Lett. 1998; 159(1):107-112. doi: 10.1111/j.1574-6968.1998.tb12848.x
Jeong EY, Sopher C, Kim IS, Lee H. Mutational study of the role of tyrosine-49 in the Saccharomyces cerevisiae xylose reductase. Yeast. 2001; 18(11):1081-1089. doi: 10.1002/yea.758
Jez JM, Bennett MJ, Schlegel BP, Lewis M, Penning TM. Comparative anatomy of the aldo-keto reductase superfamily. Biochem J. 1997; 326(3):625-636. doi: 10.1042/bj3260625
McCarthy T, Hanniffy O, Savage AV, Tuohy MG. Catalytic properties and mode of action of three endo-betaglucanases from Talaromyces emersonii on soluble beta-1,4-and beta-1,3;1,4-linked glucans. Int J Biol Macromol. 2003; 33(1-3):141-148. doi: 10.1016/S0141-8130(03)00080-1
Tuohy MG, Puls J, Claeyssens M, Vrsanská M, Coughlan MP. The xylan-degrading enzyme system of Talaromyces emersonii: novel enzymes with activity against aryl beta-D-xylosides and unsubstituted xylans. Biochem J. 1993; 290(2):515-523. doi: 10.1042/bj2900515
Tuohy MG, Walsh DJ, Murray PG, Claeyssens M, Cuffe MM, Savage AV, Coughlan MP. Kinetic parameters and mode of action of the cellobiohydrolases produced by Talaromyces emersonii. Acta Biochim Biophys. 2002; 1596(2):366-380. doi: 10.1016/s0167-4838(01)00308-9
Chang Q, Griest T, Harter T, Petrash J. Functional studies of aldo-keto reductases in Saccharomyces cerevisiae. Acta Biochim Biophys. 2007; 1773(3): 321-329. doi: 10.1016/j.bbamcr.2006.10.009
Diderich JA, Schepper M, van Hoek P, Luttik MA, van Dijken JP, Pronk JT, Klaassen P, Boelens HF, de Mattos MJ, van Dam K. Glucose uptake kinetics and transcription of HXT genes in chemostat cultures of Saccharomyces cerevisiae. Biol Chem. 1999; 274:15350-15359. doi: 10.1074/jbc.274.22.15350
Hamacher T, Becker J, Gárdonyi M, Hahn-Hägerdal B, Boles E. Characterization of the xylose-transporting properties of yeast hexose transporters and their influence on xylose utilization. Microbiology. 2002; 148(9): 2783-2788. doi: 10.1099/00221287-148-9-2783
Du J, Li S, Zhao H. Discovery and characterization of novel D-xylose-specific transporters from Neurospora crassa and Pichia stipitis. Mol Biosyst. 2010; 6(11): 2150-2156. doi: 10.1039/c0mb00007h
Leandro MJ, Goncalves P, Spencer-Martins. Two glucose/xylose transporter genes from the yeast Candida intermedia: first molecular characterization of a yeast xylose-H+ symporter. Biochem J. 2006; 395(3):543-549. doi: 10.1042/BJ20051465
Leandro MJ, Spencer-Martins, Goncalves P. The expression in Saccharomyces cerevisiae of a glucose/xylose symporter from Candida intermedia is affected by the presence of a glucose/xylose facilitator. Microbiology. 2008; 154(6):1646-1655. doi: 10.1099/mic.0.2007/015511-0
Runquist D, Fonseca C, Radstrom P, Spencer-Martins I, Hahn-Hagerdal B. Expression of the Gxf1 transporter from Candida intermedia improves fermentation performance in recombinant xylose-utilizing Saccharomyces cerevisiae. J Appl Microbiol Biotechnol. 2009; 82(1):123-130.
doi: 10.1007/s00253-008-1773-y
Watanabe S, Kodaki T, Makino K. Complete Reversal of Coenzyme Specificity of Xylitol Dehydrogenase and Increase of Thermostability by the Introduction of Structural Zinc. Biol Chem. 2005; 280(11):10340-10349. doi: 10.1074/jbc.M409443200
Jeppsson M, Johansson B, Hahn-Hägerdal B, Gorwa-Grauslund, MF. Reduced oxidative pentose phosphate pathway flux in recombinant xylose-utilizing Saccharomyces cerevisiae strains improves the ethanol yield from xylose. J Appl Environ Microbiol. 2002; 68(4):1604-1609.
doi: 10.1128/AEM.68.4.1604-1609.2002
Johansson B, Hahn-Hagerdal B. The non-oxidative pentose phosphate pathway controls the fermentation rate of xylulose but not of xylose in Saccharomyces cerevisiae TMB3001. FEMS Yeast Res. 2002; 2(3):277-282. doi: 10.1111/j.1567-1364.2002.tb00095.x
Verho R, Londesborough J, Penttila M, Richard P. Engineering redox cofactor regeneration for improved pentose fermentation in Saccharomyces cerevisiae. J Appl Environ Microbiol. 2003; 69(10):5892-5897. doi: 10.1128/AEM.69.10.5892-5897.2003
Metzger MH, Hollenberg CP. Amino acid substitutions in the yeast Pichia stipitis xylitol dehydrogenase coenzyme-binding domain affect the coenzyme specificity. Eur J Biochem. 1995; 228(1):50-54. doi: 10.1111/j.1432-1033.1995.0050o.x
Brat D, Boles E, Wiedemann B. Functional expression of a bacterial xylose isomerase in Saccharomyces cerevisiae. J Appl Environ Microbiol. 2009; 75(8):2304-2311. doi: 10.1128/AEM.02522-08
Ota M, Sakuragi H, Morisaka H, Kuroda K, Miyake H, Tamaru Y, Ueda M. Display of Clostridium cellulovorans Xylose Isomerase on the Cell Surface of Saccharomyces cerevisiae and its Direct Application to Xylose Fermentation. Biotechnol Prog. 2013; 29(2):346-351. doi:10.1002/btpr.1700
Walfridsson M, Bao X, Anderlund M, Lilius G, Bulow L, Hahn-Hagerdal B. Ethanolic fermentation of xylose with Saccharomyces cerevisiae harboring the Thermus thermophilus xylA gene, which expresses an active xylose (glucose) isomerase. J Appl Environ Microbiol. 1996; 62(12):4648-4651
Bielen A, Teparić R, Vujaklija D, Mrša V. Microbial Anchoring Systems for Cell-Surface Display of Lipolytic Enzymes. Food Technol Biotechnol. 2014; 52(1):16-34
Teparić R, Stuparević I, Mrša V. Incorporation of Homologous and Heterologous Proteins in the Saccharomyces cerevisiae Cell Wall. Food Technol Biotechnol. 2010; 48(3):317-328
Teparić R, Didak B, Ščulac E, Mrša V. Genetic immobilization of RNase Rny1p at the Saccharomyces cerevisiae cell surface. J Gen Appl Microbiol. 2013; 59(1):75-82. doi: 10.2323/jgam.59.75
Teparić R, Mrša V. Overview of systems and techniques for surface display of recombinant proteins in yeast S. cerevisiae. Appl Food Biotechnology. 2016; 3(1):3-14. doi:10.22037/afb.v3i1.9457
Kubiseski TJ, Flynn TG. Studies on Human Aldose Reductase. Probing the role of Arginine 268 by site-directed mutagenesis. Biol Chem. 1995; 270(28):16911-16917. doi: 10.1074/jbc.270.28.16911
Petschacher B, Nidetzky B. Engineering Candida tenuis xylose reductase for improved utilization of NADH: antagonistic effects of multiple side chain replacements and performance of site-directed mutants under simulated in vivo conditions. J Appl Environ Microbiol. 2005; 71(10):6390-6393. doi: 10.1128/AEM.71.10.6390-6393.2005
- Abstract Viewed: 1058 times
- PDF Downloaded: 599 times